Public Resources
Here is a curated list of publicly available tools and databases that we like to use in our projects

Intro to Coding Course Summer 2019
8 Lessons

PAVER
PAVER is an R package and R Shiny web application for summary interpretation and visualization of pathway analyses.

Allen Brain Map
The Allen Brain Atlases capture patterns of gene expression across the brain in various species

ARCHS4
ARCHS4 is a web resource that makes the majority of published RNA-seq data from human and mouse available at the gene and transcript …

cBioPortal
The cBioPortal for Cancer Genomics is an open-access, open-source resource for interactive exploration of multidimensional cancer …

Dark Kinase Knowledgebase
The Dark Kinase Knowledgebase is an online compendium of knowledge and experimental results of understudied kinases

Drug Central
DrugCentral provides information on active ingredients chemical entities, pharmaceutical products, drug mode of action, indications, …

Drug Gene Budger
Drug Gene Budger (DGB) is a web-based and mobile application developed to assist investigators in order to prioritize small molecules …

Drugmonizome
Drugmonizome is a web portal for querying sets of known drugs and computing significant biomedical terms that drugs in the set share.
Drugshot
DrugShot is a search engine that accepts any search term to return a list of small molecules that are mostly associated with the search …

Enrichr
Enrichr is a gene set search engine that enables the querying of hundreds of thousands of annotated gene sets.

Gene Expression Omnibus (GEO)
Gene Expression Omnibus (GEO) is a database repository of high throughput gene expression data and hybridization arrays, chips, …

Geneshot
Geneshot enables researchers to enter arbitrary search terms, to receive ranked lists of genes relevant to the search terms.

Genomic Data Commons Data Portal
A data portal for storing, analyzing, and sharing genomic and clinical data from patients with cancer.

Genotype-Tissue Expression (GTEx)
The Genotype-Tissue Expression (GTEx) project is an ongoing effort to build a comprehensive public resource to study tissue-specific …
GWAS Catalog
The NHGRI-EBI Catalog of human genome-wide association studies

Harmonizome
Harmonizome is a collection of information about genes and proteins from 114 datasets provided by 66 online resources.

iLINCS
iLINCS (Integrative LINCS) is an integrative web platform for analysis of LINCS data and signatures

iPTMnet
iPTMnet is a bioinformatics resource for integrated understanding of protein post-translational modifications (PTMs) in systems biology …

Kinase Enrichment Analysis 3 (KEA3)
KEA3 is a web application that infers overrepresentation of upstream kinases whose putative substrates are in a user-inputted list of …

Kinase MD
Kinase MD is kinase mutations and drug response database

Kinase Screen
Kinase Screen is a kinase profiling inhibitor database of 254 commonly used signal transduction inhibitors.

Kinase.com
Kinase.com explores the functions, evolution and diversity of protein kinases, the key controllers of cell behavior

Kinexus
Kinexus is a online resource developed by Kinexus Bioinformatics Corporation to foster the study of cell signalling systems to advance …
KinMap
KinMap is a web-based tool for interactive navigation through human kinome data

Open Targets
Open Targets is a tool that supports systematic identification and prioritisation of potential therapeutic drug targets.

Pathway Commons
Pathway Commons let you access and discover data integrated from public pathway and interactions databases

Pharos
Pharos is an integrated web-based informatics platform for the analysis of data aggregated by the Illuminating the Druggable Genome …

PhosphoSitePlus
PhosphoSitePlus provides comprehensive information and tools for the study of protein post-translational modifications (PTMs) including …

PiNET
PiNET is a versatile web platform for downstream analysis and visualization of proteomics data,

PRIDE
PRIDE is a centralized, standards compliant, public data repository for mass spectrometry proteomics data

Proteomic Data Commons (PDC)
PDC let you access highly curated and standardized cancer-related biospecimen, clinical, and proteomic data

Reactome
Reactome is an open-source, open access, manually curated and peer-reviewed pathway database.

STRING
STRING is a database of known and predicted protein-protein interactions
The Human Protein Atlas
The Human Protein Atlas aims to map all the human proteins in cells, tissues, and organs using an integration of various omics …

TIN-X
TIN-X is an interactive visualization tool for discovering interesting associations between diseases and potential drug targets.

UniProt
UniProt is a comprehensive resource for protein sequence and annotation data

X2K
X2K infers upstream regulatory networks from signatures of differentially expressed genes.

Virtual Screening Workshop
Neurosciences webinars series 2021

KRSA App
KRSA is an R package and R Shiny web application for an end-to-end upstream kinase analysis of kinome array data

CDRL Spring 2020 Webinar Series
CDRL Spring 2020 Webinar and Workshops Series
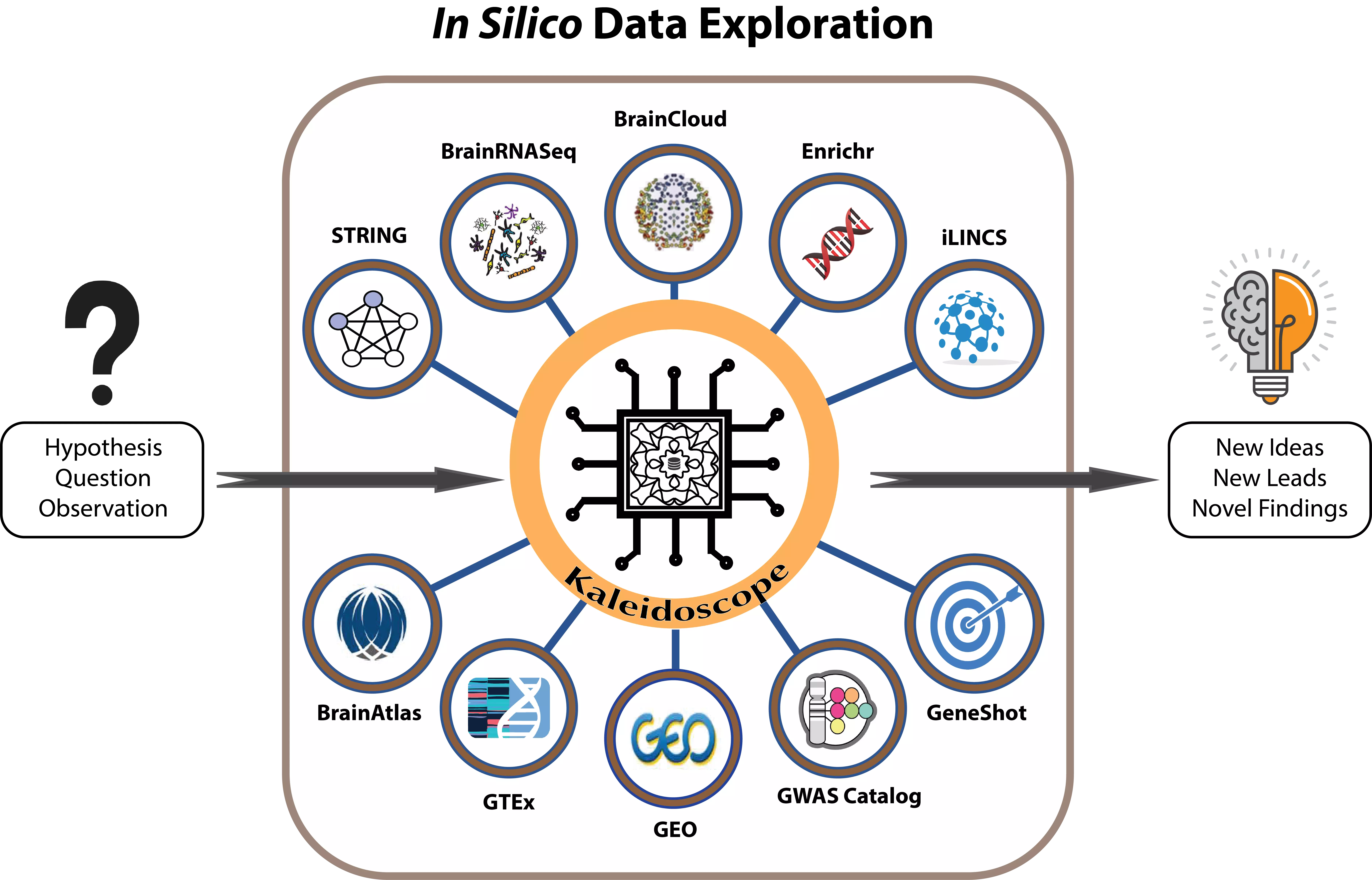
Kaleidoscope App
Kaleidoscope is a web application that provides a platform for an easy access to biological databases and bioinformatics tools

Kaleidoscope Poster (SOBP & GLBIO 2019)
Kaleidoscope Poster Presentation at the Society of Biological Psychiatry Conference & Great Lakes Bioinformatics Conference in 2019

Neurosciences Mini Symposium 2019
The University of Toledo Neurosciences Mini Symposium